November 16, 2018 feature
Solving mazes with single-molecule DNA navigators
The field of intelligent nanorobotics is based on the great promise of molecular devices with information processing capabilities. In a new study that supports the trend of DNA-based information carriers, scientists have engineered a DNA navigator system that can perform single-molecule, parallel, depth-first search operations on a two-dimensional origami platform.
Pathfinding operations with DNA navigators make use of a localized strand exchange cascade process initiated at a unique trigger site on the origami platform. Automatic progression along paths is enabled by DNA hairpins containing a universal traversal sequence. By design, each single-molecule navigator can autonomously explore any of the possible paths through a 10-vertex rooted tree constructed in the study. The mazes were equivalent to a tree with an entrance at the root and an exit through one of the leaves. The study conducted by Jie Chao and co-workers resulted in exploring all paths taken by the DNA navigators to extract a specific solution path that connected a given pair of start and end vertices in the maze. As a result, the solution path was laid plainly on the origami platform and illustrated using single-molecule imaging. The approach is now published in Nature Materials, detailing the realization of molecular materials with embedded biomolecular computational functions to operate at the level of the single-molecule with potential to engineer intelligent nanorobots for future applications in industry and medicine.
Sophisticated molecular tools were used in the past to create molecular machines that convert chemical, photonic or electric energy into rotary or linear movements at the nanoscale. For instance, Brownian motion at the nanoscale can be controllably converted into directed movements within DNA-based nanomachines using DNA hybridization reactions. Such DNA-based machines operate autonomously by following an embedded 'molecular program' pre-designed as a cascade reaction manually triggered via an external stimulus for each step of the operation.
The focus of the field has progressively shifted to actualize DNA-based logic circuits using aptamers and DNAzymes to design molecular logic gates. For example, in 2006, Stojanovic and co-workers integrated more than 100 DNA logic gates to engineer an automation calledMAYA-II to play a game of Tic-Tac-Toe. Preceding studies demonstrated an enzyme free computing system based on hybridization chain reactions (HCR) to create logic-gates and logic circuits for more robust and efficient performance than the original systems.
In the present study by Chao et al, the same basic principle of the HCR reaction scheme was used in a different computational context to develop a single-molecule DNA-navigator system. The platform explored all possible paths through a tree graph designed on an origami structure as a simply connected maze without cyclic paths. Such DNA origami structures are information-bearing nanostructures by nature with well-defined nanoscale geometry. The maze could be explored by proximal strand exchange cascade (PSEC) based on hybridization chain reactions. The researchers demonstrated that a system with a large number of single-molecule DNA navigators could collectively conduct parallel depth-first search (PDFS) on the tree to efficiently perform maze solving within 2-D origami. Initially the researchers conducted studies to test the PSEC design.
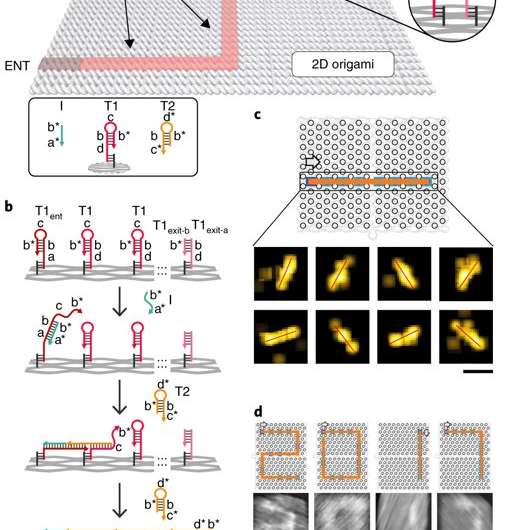
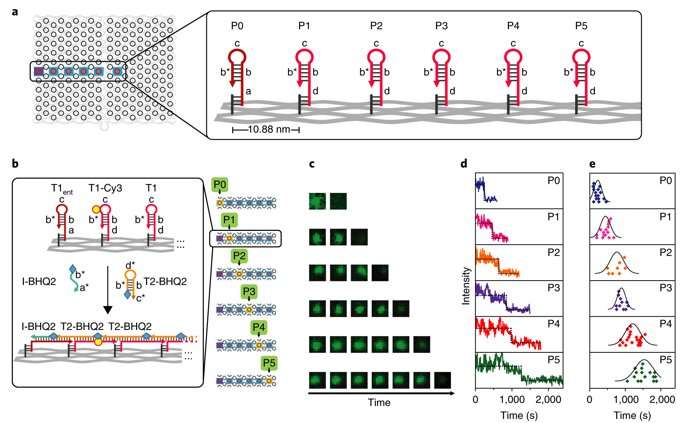
The proximal strand exchange cascade (PSEC) system (working principle of the DNA navigator) was facilitated on a rectangular origami substrate made of three components, which included the physical implementation of a tree graph, full strands and an initiator strand. Vacant areas without staple extensions corresponded to walls in the maze, preventing propagation of the strand exchange cascade. The entrance and exit were defined and denoted as ENT and EXIT respectively. In the second component, two types of DNA hairpins, T1 and T2, were used as fuels to drive the PSEC on the tree graph. The two hairpins coexisted metastably in solution to hybridize and fuel the PSEC process with free energy .
By design, information only propagated through the network in the presence of an initiator (Initiator I). Upon addition of initiator I, PSEC was conducted and observed using atomic force microscopy (AFM). To visualize an established formation, the scientists enabled DNA-navigator-based formation of the number 2017 as a proof-of-principle. Another technique known as DNA-PAINT was employed as a single-molecule, super-resolution imaging technique to reveal molecular features at the nanoscale to further substantiate the PSEC-based path paving process. The on-origami PSEC was highly specific, without intra- or inter-origami crosstalk.
Kinetics of the process were investigated at the single-molecule level in the study using time-resolved total internal reflection fluorescence microscopy (TIRF) in a setup with a prescribed starting point (P0) and five intermediate steps (P1-P5). Fluorescence in the setup was quenched using fluorescence resonance energy transfer (FRET), and the cascade was observed in real time by recording fluorescence signals continuously. The average speed of propagation was recorded to be 2.46 nm per minute, propagation across the straight line (54.4 nm) took approximately 22 minutes.
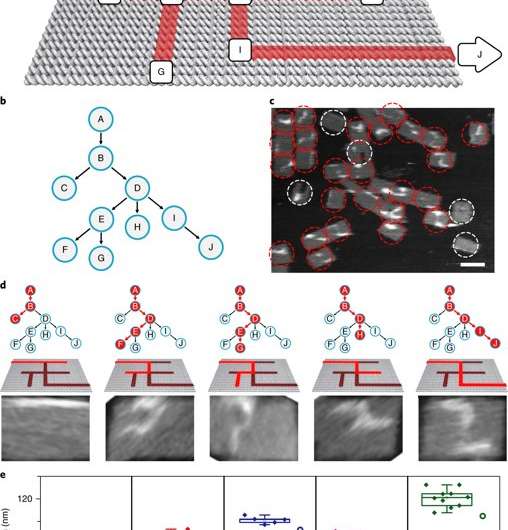
The scientists then constructed the main model maze with 10 vertices that included an entrance vertex A and an exit vertex J, three junctions (B, D, E) an intermediate vertex (I) and four dead ends (C, F, G, H). Each path of the maze that was equivalent to a tree with 10 vertices was investigated using DNA investigators starting at root A. The PSEC reactions produced a mixture of various paths on the maze, confirmed with AFM. Each individual PSEC could progress on one of the five possible paths. Statistical analysis of the length distribution showed that the measured paths coincided well with the values predicted.
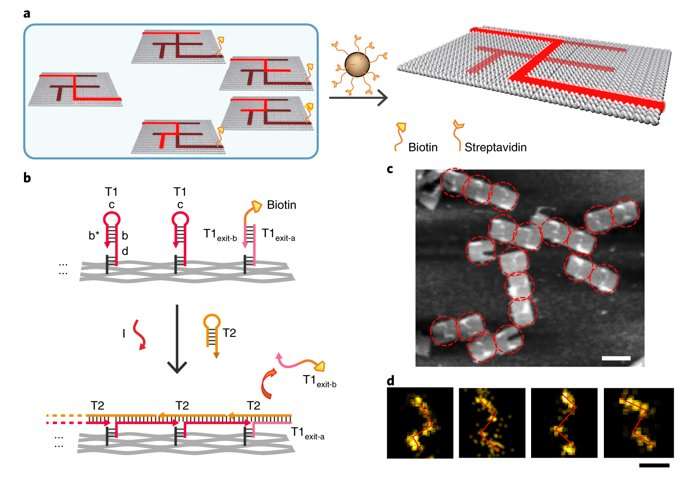
To prevent the navigators from propagating through a wrong path with dead ends, the scientists designed a streptavidin-biotin tag-based method to selectively eliminate inaccurate path navigation. Only the correct path (PABDIJ) was followed in the maze therefore. The computational context used in the study allowed exploration of paths through tree graphs defined on the origami. The autonomous path explored by the DNA navigators proceeded unidirectionally and irreversibly, turning at junctions and corners on the origami platform as they were designed to. The design enabled parallel depth first search (PDFS) allowing each DNA navigator to individually explore any one of the paths through the given graph at a defined speed, greater than that previously achieved.
The main advantage of the described biomolecular computer schemes in comparison to conventional electronic computing is they can be interfaced directly with biologically relevant processes. As a result, the scientists envision translational biomedical sensing and decision-making platforms with DNA origami and single-molecule diagnostics using decision trees. Future applications will also include simple sensors or those coupled to a molecular actuator to trigger downstream molecular cascades.
More information: Jie Chao et al. Solving mazes with single-molecule DNA navigators, Nature Materials (2018). DOI: 10.1038/s41563-018-0205-3
Renjun Pei et al. Training a molecular automaton to play a game, Nature Nanotechnology (2010). DOI: 10.1038/nnano.2010.194
S. M. Douglas et al. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads, Science (2012). DOI: 10.1126/science.1214081
Journal information: Nature Materials , Nature Nanotechnology , Science
© 2018 Science X Network