New targets in the battle against antibiotic resistance

Bacteria are increasingly resistant to available antibiotics. A team of chemists from the Technical University of Munich (TUM) have now identified important enzymes in the metabolism of staphylococci. Blocking these enzymes in a targeted manner starves them.
"Medicine needs new weapons against bacteria," says Prof. Stephan Sieber. "Many bacteria are already resistant to common drugs. An important goal of the research is thus to identify new points of attack."
Together with Ph.D. student Annabelle Hoegl and his team at the Chair of Organic Chemistry II at the Technical University of Munich, the researcher has developed a procedure for isolating and metabolizing enzymes that control metabolism. "If we could block these enzymes," Stephan Sieber explains the goal, "we could more or less starve out the pathogens."
The new methodology was tested on Staphylococcus aureus. The bacterium is widespread, with many species resistant to antibiotics. Staphylococci comprise thousands of proteins. "Isolating enzymes with specific properties from this haystack, identifying them and investigating their function presented a real challenge," recalls Sieber.
The enzymes targeted by the research team use vitamin B6 to accelerate chemical reactions in the cell. A crucial component of vitamin B6 is pyridoxal phosphate, or PLP. Without the PLP-dependent enzymes, the bacteria's metabolism would come to a halt, starving the microorganisms to death.
Tracking down enzymes with markers
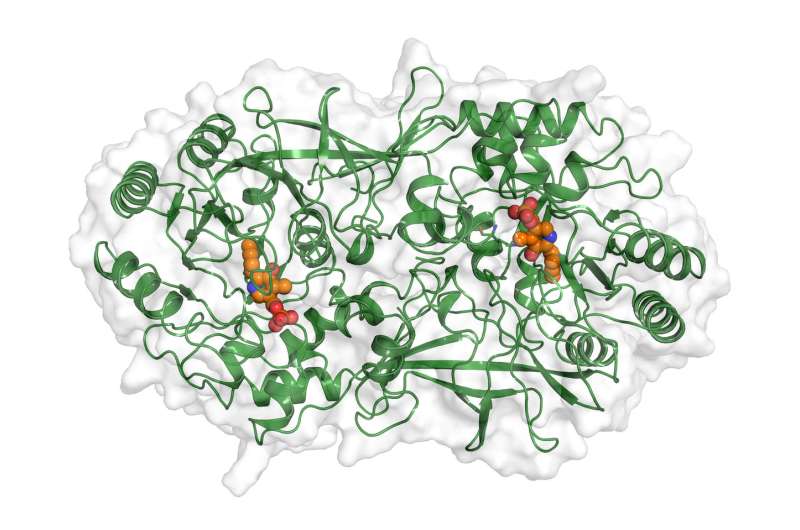
The team used a chemically modified pyridoxal phosphate to detect such PLP-dependent enzymes. The marked molecules were added to a nutrient solution in which the Staphylococcus aureus bacteria proliferate.
Since the solution contained no natural PLP, the PLP-dependent enzymes incorporated the markers, using them for metabolism. The chemists then ruptured the bacteria using ultrasound and fished out the enzymes carrying the markers.
The principle of molecular fishing, or "protein profiling," as it is called, is not completely new. But, the TUM scientists are the first to utilize this methodology to investigate PLP-dependent enzymes.
"We were able to demonstrate that the method is very efficient," emphasizes Sieber. "Many physiologically important enzymes in Staphylococcus depend on pyridoxal phosphate. We isolated 73% of these enzymes, analyzed them using mass spectrometry and identified them."
In addition, the team uncovered previously unknown PLP-dependent enzymes and deciphered their functions. "Therewith we discovered an untapped treasure in our search for new antibiotic targets," says the chemist.
This insight can now be used to develop new active agents against bacteria. In the next step, the researchers hope to investigate the functions of the enzymes in greater detail and determine how the metabolism of bacteria can be blocked in a targeted manner without damaging healthy human cells.
More information: Annabelle Hoegl et al, Mining the cellular inventory of pyridoxal phosphate-dependent enzymes with functionalized cofactor mimics, Nature Chemistry (2018). DOI: 10.1038/s41557-018-0144-2
Journal information: Nature Chemistry
Provided by Technical University Munich