Combining microbial and chemical fingerprints for forensics applications
Researchers at the UC San Diego School of Medicine and Center for Microbiome Innovation have for the first time combined technologies that detect the presence of microbes and chemicals to identify "who touched what" in a manmade space.
The new method fills a gap in current forensic technologies, and could have a variety of applications, including criminal profiling and environmental exposure studies. The study was published recently in Scientific Reports.
Cliff Kapono, famous for the Surfer Biome Project and visiting oceans around the world on a surfboard made of algae, is studying human-environment interactions in the lab of UC San Diego School of Medicine and Skaggs School of Pharmacy and Pharmaceutical Sciences professor Pieter Dorrestein. The Dorrestein lab has conducted studies on what small molecules left on surfaces reveal about one's personal lifestyle in the past—including a study that looked at molecules on phone screens, and one that pinpointed coffee spills in an office by detecting trace amounts of caffeine.
The team again visited the office for this study—but this time, they went a step further: the researchers looked to see whether they could identify the individuals inhabiting the space and which items they touched in the office by identifying the microbes and chemicals present on their bodies.
"We found that we can detect molecules that relate to an individual's diet and behavior, which includes personal hygiene, medication and lifestyle," said Kapono, who is the first author on the paper. "We also showed that an individual's microbiome and metabolome are unique, and because of that, we can track who touched what in the room."
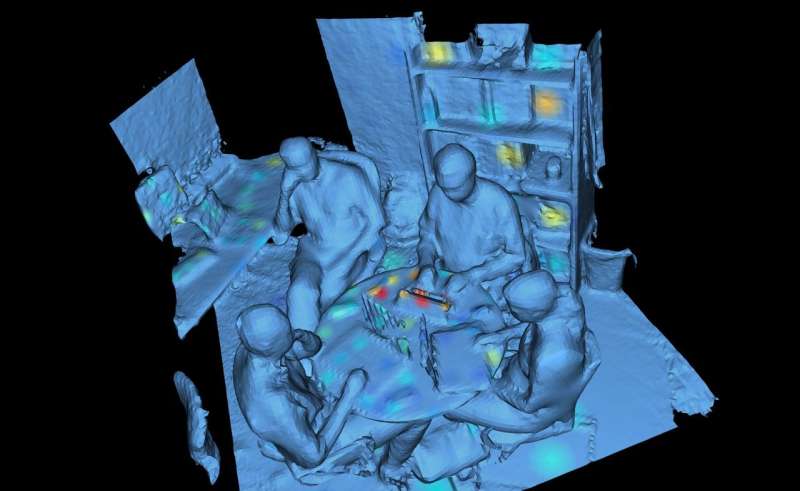
The office environment included a desk, bookshelf, conference table, four chairs, garbage and recycling cans, the carpet-covered floor, phones, computers and computer accessories and walls. The team swabbed nearly 400 sites inside the office, and also took samples from four volunteers who frequented the space.
"The main inhabitant of the office was Volunteer 3," said Kapono. "Volunteer 1 had only been in the office twice in the two years prior to sampling, and volunteers 2 and 4 frequented the office 2–4 times a month for short periods of time during each visit."
The team found DEET insect repellent and marine bacteria on volunteer 3, which suggested that this person was an outdoorsman. A common pesticide was found on the computer, hands, and phone of volunteer 2.
Using statistical analysis, the researchers determined the uniqueness of each individual in terms of their microbiota and chemicals present on their skin, and used a bioinformatics tool to track who touched what.
The combination of technologies used to detect the presence of microbes and small molecules to identify the volunteers and determine who touched what highlights the innovative nature of microbiome research on campus. The UC San Diego Center for Microbiome Innovation, led by world-renowned microbiome researcher and professor of pediatrics and computer science and engineering Rob Knight, was created in 2016 as part of UC San Diego Jacobs School of Engineering Dean Albert P. Pisano's agile research centers initiative. Funds for the establishment of the Center came in part from UC San Diego Chancellor Pradeep K. Khosla's Microbiome and Microbial Sciences Initiative.
Since it launched, the Center has brought in upwards of $2 million in external funding and grown to 120+ interdisciplinary faculty members with expertise ranging from medicine to big data analysis. Recent media coverage has highlighted some of them: for example, Richard Gallo's team recently identified a strain of Staphylococcus epidermidis, common on healthy human skin, that exerts a selective ability to inhibit the growth of some cancers. In addition, some of the world's largest citizen science projects and collaborative scientific studies—including the American Gut Project and the Earth Microbiome Project—are run out of the Knight lab.
Together with industry, Center members are accelerating microbiome research through the development of new tools, such as the 3-D data visualization tool used in this study.
"Ili"
"Before we analyzed the samples, we created a 3-D model of the room using a 3-D scanner on an iPad," said Kapono. "We then used an open-source tool we developed called 'ili to map the data onto the model. Anybody can do this—that's the point. Often, we just look at graphs. We wanted to enable people to visualize data—doesn't matter what kind—in 3-D."
"There are many ways that investigators try to answer the question, 'Who was where?'—including through DNA analysis, fingerprinting, and by talking to witnesses," said Kapono. By combining the detection of microbial and chemical signatures, we've created another method that can detect trace amounts of molecules—not all forensics tools can do that. It's really about checks and balances."
But, this isn't just meant to put people in jail, said Kapono. "We were able to detect soil and ocean bacteria in the office. This highlights the fact that we bring nature home with us or to the places we work. For that reason, this technology will be very interesting to institutions and individuals whose entire day revolves around a built environment. We unintentionally exchange molecular signatures with everyone around us and the spaces we occupy. If companies can link molecules or bacteria to a safer, better work environment, then perhaps they can implement the necessary changes to improve their workforce."
These data also support Kapono's Surfer Biome Project, which looks to explore how humans interact with the natural world on a molecular level. The Surfer Biome Project is expected to be published Summer 2018.
More information: Clifford A. Kapono et al. Creating a 3D microbial and chemical snapshot of a human habitat, Scientific Reports (2018). DOI: 10.1038/s41598-018-21541-4
Journal information: Scientific Reports
Provided by University of California - San Diego